Assemblies
Assembly Name:
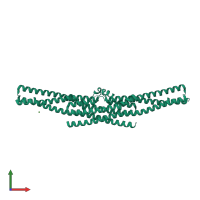
Endophilin-A1
Multimeric state:
homo dimer
Accessible surface area:
23628.37 Å2
Buried surface area:
5339.67 Å2
Dissociation area:
2,632.71
Å2
Dissociation energy (ΔGdiss):
44.98
kcal/mol
Dissociation entropy (TΔSdiss):
13.97
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-189292
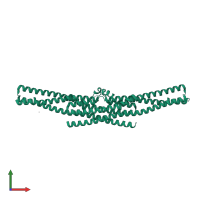
Assembly Name:
Endophilin-A1
Multimeric state:
homo dimer
Accessible surface area:
22588.76 Å2
Buried surface area:
5141.07 Å2
Dissociation area:
2,570.53
Å2
Dissociation energy (ΔGdiss):
44.63
kcal/mol
Dissociation entropy (TΔSdiss):
13.84
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-189292
Macromolecules
Chains: A, B, C, D
Length: 256 amino acids
Theoretical weight: 29.06 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: BAR domain
InterPro:
CATH: Arfaptin homology (AH) domain/BAR domain
SCOP: BAR domain
Length: 256 amino acids
Theoretical weight: 29.06 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q99962 (Residues: 1-154, 155-247; Coverage: 70%)
Q99962 (Residues: 1-154, 155-247; Coverage: 70%)
Pfam: BAR domain
InterPro:
CATH: Arfaptin homology (AH) domain/BAR domain
SCOP: BAR domain