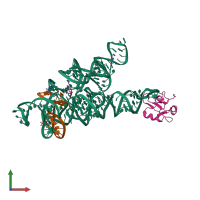
Assemblies
Assembly Name:
U1 small nuclear ribonucleoprotein A and RNA
Multimeric state:
hetero tetramer
Accessible surface area:
36813.09 Å2
Buried surface area:
8889.89 Å2
Dissociation area:
898.76
Å2
Dissociation energy (ΔGdiss):
4.9
kcal/mol
Dissociation entropy (TΔSdiss):
11.81
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-140476
Macromolecules
Chain: A
Length: 98 amino acids
Theoretical weight: 11.34 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: RNA recognition motif
InterPro:
SCOP: Canonical RBD
Length: 98 amino acids
Theoretical weight: 11.34 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P09012 (Residues: 1-98; Coverage: 35%)
P09012 (Residues: 1-98; Coverage: 35%)
Pfam: RNA recognition motif
InterPro:
- Nucleotide-binding alpha-beta plait domain superfamily
- RNA-binding domain superfamily
- U1 small nuclear ribonucleoprotein A, RNA recognition motif 1
- RNA-binding region-containing protein RBM41/RNPC3
- RNA recognition motif domain
SCOP: Canonical RBD
Name:
197-MER
Representative chains: B
Source organism: Homo sapiens [9606]
Expression system: Not provided
Length: 197 nucleotides
Theoretical weight: 64.06 KDa
Representative chains: B
Source organism: Homo sapiens [9606]
Expression system: Not provided
Length: 197 nucleotides
Theoretical weight: 64.06 KDa
Name:
5'-R(*AP*AP*GP*CP*CP*AP*CP*AP*CP*AP*AP*AP*CP*CP*AP*GP*AP*CP*GP*GP*CP*C)-3'
Representative chains: C
Source organism: Homo sapiens [9606]
Expression system: Not provided
Length: 22 nucleotides
Theoretical weight: 7.05 KDa
Representative chains: C
Source organism: Homo sapiens [9606]
Expression system: Not provided
Length: 22 nucleotides
Theoretical weight: 7.05 KDa
Name:
5'-R(*CP*AP*(5MU))-3'
Representative chains: D
Source organism: Homo sapiens [9606]
Expression system: Not provided
Length: 3 nucleotides
Theoretical weight: 910 Da
Representative chains: D
Source organism: Homo sapiens [9606]
Expression system: Not provided
Length: 3 nucleotides
Theoretical weight: 910 Da