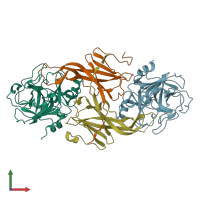
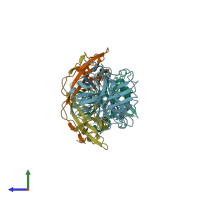
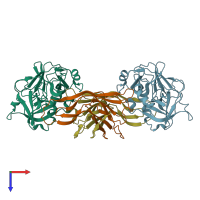
Assemblies
Assembly Name:
Coagulation factor XIa light chain and Ecotin
Multimeric state:
hetero tetramer
Accessible surface area:
33787.25 Å2
Buried surface area:
9775.71 Å2
Dissociation area:
2,785.69
Å2
Dissociation energy (ΔGdiss):
18.91
kcal/mol
Dissociation entropy (TΔSdiss):
14.64
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-137288
Macromolecules
Chains: A, B
Length: 238 amino acids
Theoretical weight: 26.83 KDa
Source organism: Homo sapiens
Expression system: Komagataella pastoris
UniProt:
Pfam: Trypsin
InterPro:
SCOP: Eukaryotic proteases
Length: 238 amino acids
Theoretical weight: 26.83 KDa
Source organism: Homo sapiens
Expression system: Komagataella pastoris
UniProt:
- Canonical:
 P03951 (Residues: 388-625; Coverage: 39%)
P03951 (Residues: 388-625; Coverage: 39%)
Pfam: Trypsin
InterPro:
- Serine proteases, trypsin domain
- Peptidase S1, PA clan
- Peptidase S1, PA clan, chymotrypsin-like fold
- Peptidase S1A, chymotrypsin family
- Serine proteases, trypsin family, histidine active site
- Serine proteases, trypsin family, serine active site
SCOP: Eukaryotic proteases
Chains: C, D
Length: 142 amino acids
Theoretical weight: 16.24 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli BL21
UniProt:
Pfam: Ecotin
InterPro:
SCOP: Ecotin, trypsin inhibitor
Length: 142 amino acids
Theoretical weight: 16.24 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli BL21
UniProt:
- Canonical:
 P23827 (Residues: 21-162; Coverage: 100%)
P23827 (Residues: 21-162; Coverage: 100%)
Pfam: Ecotin
InterPro:
- Proteinase inhibitor I11, ecotin
- Proteinase inhibitor I11, ecotin, gammaproteobacteria
- Ecotin superfamily
- Ecotin, C-terminal
SCOP: Ecotin, trypsin inhibitor