Assemblies
Assembly Name:
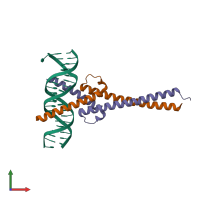
MYC-MAX transcriptional activator complex and DNA
Multimeric state:
hetero tetramer
Accessible surface area:
16649.09 Å2
Buried surface area:
7958.13 Å2
Dissociation area:
1,481.43
Å2
Dissociation energy (ΔGdiss):
32.43
kcal/mol
Dissociation entropy (TΔSdiss):
11.95
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-114266
Assembly Name:
MYC-MAX transcriptional activator complex and DNA
Multimeric state:
hetero tetramer
Accessible surface area:
16266.81 Å2
Buried surface area:
7685.65 Å2
Dissociation area:
1,423.63
Å2
Dissociation energy (ΔGdiss):
27.46
kcal/mol
Dissociation entropy (TΔSdiss):
11.88
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-114266
Macromolecules
Chains: A, D
Length: 88 amino acids
Theoretical weight: 10.47 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam:
InterPro:
SCOP: HLH, helix-loop-helix DNA-binding domain
Length: 88 amino acids
Theoretical weight: 10.47 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P01106 (Residues: 368-449; Coverage: 18%)
P01106 (Residues: 368-449; Coverage: 18%)
Pfam:
InterPro:
- Helix-loop-helix DNA-binding domain superfamily
- Myc-type, basic helix-loop-helix (bHLH) domain
- Transcription regulator Myc
- Leucine zipper, Myc
SCOP: HLH, helix-loop-helix DNA-binding domain
Chains: B, E
Length: 83 amino acids
Theoretical weight: 9.8 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Helix-loop-helix DNA-binding domain
InterPro:
CATH: Helix-loop-helix DNA-binding domain
SCOP: HLH, helix-loop-helix DNA-binding domain
Length: 83 amino acids
Theoretical weight: 9.8 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P61244 (Residues: 23-102; Coverage: 50%)
P61244 (Residues: 23-102; Coverage: 50%)
Pfam: Helix-loop-helix DNA-binding domain
InterPro:
CATH: Helix-loop-helix DNA-binding domain
SCOP: HLH, helix-loop-helix DNA-binding domain
Name:
5'-D(*CP*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3'
Representative chains: F, G, H, J
Length: 19 nucleotides
Theoretical weight: 5.81 KDa
Representative chains: F, G, H, J
Length: 19 nucleotides
Theoretical weight: 5.81 KDa