EC 1.3.1.98: UDP-N-acetylmuramate dehydrogenase
Reaction catalysed:
UDP-N-acetyl-alpha-D-muramate + NADP(+) = UDP-N-acetyl-3-O-(1-carboxyvinyl)-alpha-D-glucosamine + NADPH
Systematic name:
UDP-N-acetyl-alpha-D-muramate:NADP(+) oxidoreductase
Alternative Name(s):
- MurB reductase
- UDP-GlcNAc-enoylpyruvate reductase
- UDP-N-acetylenolpyruvoylglucosamine reductase
- UDP-N-acetylglucosamine-enoylpyruvate reductase
- Uridine diphospho-N-acetylglucosamine-enolpyruvate reductase
- Uridine diphosphoacetylpyruvoylglucosamine reductase
- Uridine-5'-diphospho-N-acetyl-2-amino-2-deoxy-3-O-lactylglucose:NADP-oxidoreductase
GO terms
Biochemical function:
Biological process:
Cellular component:
Sequence families
 Protein families (Pfam)
Protein families (Pfam)
 InterPro annotations
InterPro annotations
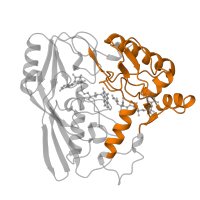
IPR036635
Domain description:
UDP-N-acetylenolpyruvoylglucosamine reductase, C-terminal domain superfamily
Occurring in:
Occurring in:
IPR011601
Domain description:
UDP-N-acetylenolpyruvoylglucosamine reductase, C-terminal
Occurring in:
Occurring in:
Structure domains
 CATH domains
CATH domains
3.90.78.10
Class:
Alpha Beta
Architecture:
Alpha-Beta Complex
Topology:
Uridine Diphospho-n-acetylenolpyruvylglucosamine Reductase; domain 1
Homology:
UDP-N-acetylenolpyruvoylglucosamine reductase, C-terminal domain
Occurring in:

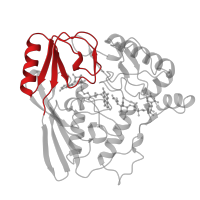
3.30.465.10
Class:
Alpha Beta
Architecture:
2-Layer Sandwich
Topology:
Uridine Diphospho-n-acetylenolpyruvylglucosamine Reductase; domain 3
Homology:
Uridine Diphospho-n-acetylenolpyruvylglucosamine Reductase; domain 3
Occurring in:

 SCOP annotations
SCOP annotations
56195
Class:
Alpha and beta proteins (a+b)
Fold:
Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain
Superfamily:
Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain
Occurring in:

56184
Class:
Alpha and beta proteins (a+b)
Fold:
FAD-binding/transporter-associated domain-like
Superfamily:
FAD-binding/transporter-associated domain-like
Occurring in: