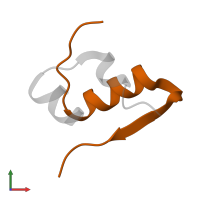Assemblies
Assembly Name:
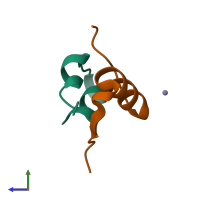
Insulin B chain
Multimeric state:
hetero dimer
Accessible surface area:
3656.55 Å2
Buried surface area:
1771.92 Å2
Dissociation area:
850.14
Å2
Dissociation energy (ΔGdiss):
11.18
kcal/mol
Dissociation entropy (TΔSdiss):
8.21
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-134237
Assembly Name:
Insulin B chain
Multimeric state:
hetero dimer
Accessible surface area:
3525.16 Å2
Buried surface area:
1901.54 Å2
Dissociation area:
917.1
Å2
Dissociation energy (ΔGdiss):
12.14
kcal/mol
Dissociation entropy (TΔSdiss):
8.2
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-134237
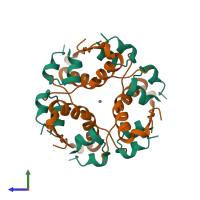
Assembly Name:
Insulin B chain
Multimeric state:
hetero dodecamer
Accessible surface area:
12137.89 Å2
Buried surface area:
20032.92 Å2
Dissociation area:
4,494.28
Å2
Dissociation energy (ΔGdiss):
65.37
kcal/mol
Dissociation entropy (TΔSdiss):
12.88
kcal/mol
Symmetry number:
6
PDBe Complex ID:
PDB-CPX-134248
Assembly Name:
Insulin B chain
Multimeric state:
hetero dodecamer
Accessible surface area:
18375.96 Å2
Buried surface area:
13794.84 Å2
Dissociation area:
1,196.48
Å2
Dissociation energy (ΔGdiss):
0.8
kcal/mol
Dissociation entropy (TΔSdiss):
11.97
kcal/mol
Symmetry number:
6
PDBe Complex ID:
PDB-CPX-134248
Assembly Name:
Insulin B chain
Multimeric state:
hetero hexamer
Accessible surface area:
10578.73 Å2
Buried surface area:
5505.79 Å2
Dissociation area:
205.48
Å2
Dissociation energy (ΔGdiss):
20.18
kcal/mol
Dissociation entropy (TΔSdiss):
19.07
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-134245
Assembly Name:
Insulin B chain
Multimeric state:
hetero hexamer
Accessible surface area:
10180.48 Å2
Buried surface area:
5905.81 Å2
Dissociation area:
204.67
Å2
Dissociation energy (ΔGdiss):
21.54
kcal/mol
Dissociation entropy (TΔSdiss):
18.97
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-134245
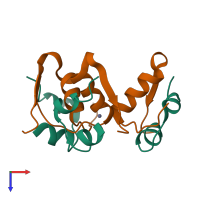
Assembly Name:
Insulin B chain
Multimeric state:
hetero tetramer
Accessible surface area:
5669.99 Å2
Buried surface area:
5188.12 Å2
Dissociation area:
813.64
Å2
Dissociation energy (ΔGdiss):
5.3
kcal/mol
Dissociation entropy (TΔSdiss):
9.66
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-134242
Assembly Name:
Insulin B chain
Multimeric state:
hetero tetramer
Accessible surface area:
5815.77 Å2
Buried surface area:
5039.41 Å2
Dissociation area:
686.05
Å2
Dissociation energy (ΔGdiss):
1.39
kcal/mol
Dissociation entropy (TΔSdiss):
9.65
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-134242
Macromolecules
Chains: A, C
Length: 21 amino acids
Theoretical weight: 2.38 KDa
Source organism: Sus scrofa
Expression system: Not provided
UniProt:
InterPro:
SCOP: Insulin-like
Length: 21 amino acids
Theoretical weight: 2.38 KDa
Source organism: Sus scrofa
Expression system: Not provided
UniProt:
- Canonical:
 P01315 (Residues: 88-108; Coverage: 25%)
P01315 (Residues: 88-108; Coverage: 25%)
InterPro:
SCOP: Insulin-like
Chains: B, D
Length: 30 amino acids
Theoretical weight: 3.43 KDa
Source organism: Sus scrofa
Expression system: Not provided
UniProt:
Pfam: Insulin/IGF/Relaxin family
InterPro:
SCOP: Insulin-like
Length: 30 amino acids
Theoretical weight: 3.43 KDa
Source organism: Sus scrofa
Expression system: Not provided
UniProt:
- Canonical:
 P01315 (Residues: 25-53; Coverage: 35%)
P01315 (Residues: 25-53; Coverage: 35%)
Pfam: Insulin/IGF/Relaxin family
InterPro:
SCOP: Insulin-like