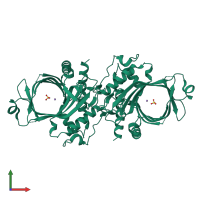
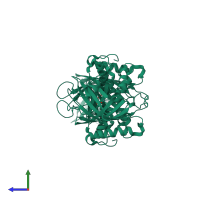
Assemblies
Assembly Name:
mRNA-capping enzyme subunit beta
Multimeric state:
homo dimer
Accessible surface area:
27651.9 Å2
Buried surface area:
5143.85 Å2
Dissociation area:
2,146.53
Å2
Dissociation energy (ΔGdiss):
24.26
kcal/mol
Dissociation entropy (TΔSdiss):
13.88
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-126743
Assembly Name:
mRNA-capping enzyme subunit beta
Multimeric state:
homo dimer
Accessible surface area:
27670.65 Å2
Buried surface area:
5208.94 Å2
Dissociation area:
2,175.13
Å2
Dissociation energy (ΔGdiss):
23.16
kcal/mol
Dissociation entropy (TΔSdiss):
13.88
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-126743
Macromolecules
Chains: A, B, C
Length: 311 amino acids
Theoretical weight: 35.69 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: mRNA capping enzyme, beta chain
InterPro:
SCOP: mRNA triphosphatase CET1
Length: 311 amino acids
Theoretical weight: 35.69 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 O13297 (Residues: 241-549; Coverage: 56%)
O13297 (Residues: 241-549; Coverage: 56%)
Pfam: mRNA capping enzyme, beta chain
InterPro:
- mRNA triphosphatase Cet1-like superfamily
- CYTH-like domain superfamily
- RNA 5'-triphosphatase Cet1/Ctl1
- mRNA triphosphatase Cet1-like
SCOP: mRNA triphosphatase CET1