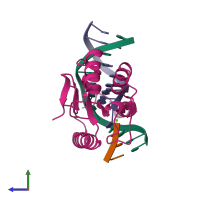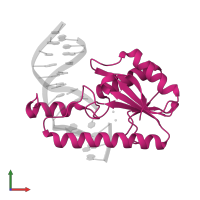Assemblies
Assembly Name:
DNA mismatch endonuclease Vsr and DNA
Multimeric state:
hetero tetramer
Accessible surface area:
10375.61 Å2
Buried surface area:
5110.4 Å2
Dissociation area:
1,848.41
Å2
Dissociation energy (ΔGdiss):
34.32
kcal/mol
Dissociation entropy (TΔSdiss):
11.23
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-113561
Macromolecules
Chain: A
Length: 155 amino acids
Theoretical weight: 17.91 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
Pfam: DNA mismatch endonuclease Vsr
InterPro:
CATH: VSR Endonuclease
SCOP: Very short patch repair (VSR) endonuclease
Length: 155 amino acids
Theoretical weight: 17.91 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
- Canonical:
 P09184 (Residues: 2-156; Coverage: 99%)
P09184 (Residues: 2-156; Coverage: 99%)
Pfam: DNA mismatch endonuclease Vsr
InterPro:
CATH: VSR Endonuclease
SCOP: Very short patch repair (VSR) endonuclease
Name:
DNA (5'-D(*AP*CP*GP*TP*AP*CP*CP*TP*GP*GP*CP*T)-3')
Representative chains: M
Source organism: Escherichia coli [562]
Expression system: Not provided
Length: 12 nucleotides
Theoretical weight: 3.64 KDa
Representative chains: M
Source organism: Escherichia coli [562]
Expression system: Not provided
Length: 12 nucleotides
Theoretical weight: 3.64 KDa
Name:
DNA (5'-D(*AP*GP*C)-3')
Representative chains: N
Source organism: Escherichia coli [562]
Expression system: Not provided
Length: 3 nucleotides
Theoretical weight: 887 Da
Representative chains: N
Source organism: Escherichia coli [562]
Expression system: Not provided
Length: 3 nucleotides
Theoretical weight: 887 Da
Name:
DNA (5'-D(P*TP*AP*GP*GP*TP*AP*CP*GP*T)-3')
Representative chains: O
Source organism: Escherichia coli [562]
Expression system: Not provided
Length: 9 nucleotides
Theoretical weight: 2.77 KDa
Representative chains: O
Source organism: Escherichia coli [562]
Expression system: Not provided
Length: 9 nucleotides
Theoretical weight: 2.77 KDa