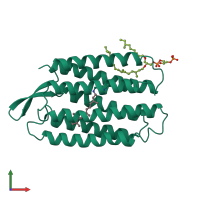
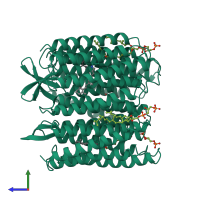
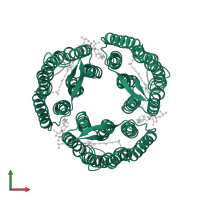
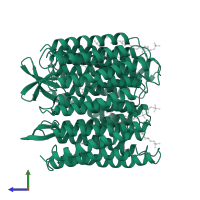
Assemblies
Assembly Name:
Bacteriorhodopsin
Multimeric state:
homo trimer
Accessible surface area:
26667.3 Å2
Buried surface area:
11089.15 Å2
Dissociation area:
3,295.11
Å2
Dissociation energy (ΔGdiss):
35.12
kcal/mol
Dissociation entropy (TΔSdiss):
26.83
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-136413
Macromolecules
Chain: A
Length: 248 amino acids
Theoretical weight: 26.8 KDa
Source organism: Halobacterium salinarum
UniProt:
Pfam: Bacteriorhodopsin-like protein
InterPro:
CATH: Rhodopsin 7-helix transmembrane proteins
SCOP: Bacteriorhodopsin-like
Length: 248 amino acids
Theoretical weight: 26.8 KDa
Source organism: Halobacterium salinarum
UniProt:
- Canonical:
 P02945 (Residues: 14-261; Coverage: 95%)
P02945 (Residues: 14-261; Coverage: 95%)
Pfam: Bacteriorhodopsin-like protein
InterPro:
CATH: Rhodopsin 7-helix transmembrane proteins
SCOP: Bacteriorhodopsin-like