EMD-8473
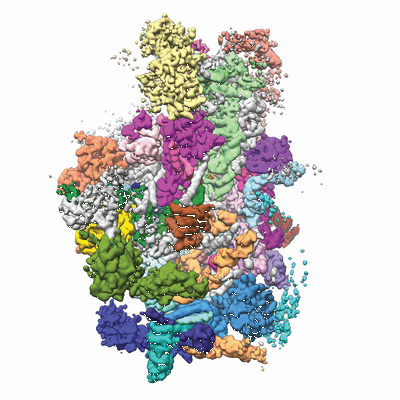
Architecture of the yeast small subunit processome
EMD-8473
Single-particle5.1 Å
 Deposition: 22/11/2016
Deposition: 22/11/2016Map released: 21/12/2016
Last modified: 13/03/2024
Concentration: 0.15
mg/mL
Buffer
pH: 7.7
Buffer components [5]:
Buffer components [5]:
| Name | Formula | Concentration | ChEBI |
|---|---|---|---|
| Sodium chloride | NaCl | 150.0 mM | |
| Tris | (HOCH2)3CNH2 | 50.0 mM | |
| EDTA | - | 1.0 mM | - |
| Biotin | - | 5.0 mM | - |
| Triton X-100 | - | 0.03 % | - |
Grid
Vitrification
Cryogen name: ETHANE
Chamber humidity: 100%
Chamber temperature: 295 K
Instrument: FEI VITROBOT MARK IV
Details: Incubated 10 seconds on the grid before blotting (blot time 2.0 seconds, blot force 0) and freezing..
Chamber humidity: 100%
Chamber temperature: 295 K
Instrument: FEI VITROBOT MARK IV
Details: Incubated 10 seconds on the grid before blotting (blot time 2.0 seconds, blot force 0) and freezing..
Microscope: FEI TITAN KRIOS
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 300 kV
C2 aperture diameter: 70.0 µm
Nominal CS: 2.7 mm
Nominal defocus: 0.6 µm - 2.6 µm
Nominal magnification: 22500.0
Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER
Cooling holder cryogen: NITROGEN
Alignment procedure: BASIC
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 300 kV
C2 aperture diameter: 70.0 µm
Nominal CS: 2.7 mm
Nominal defocus: 0.6 µm - 2.6 µm
Nominal magnification: 22500.0
Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER
Cooling holder cryogen: NITROGEN
Alignment procedure: BASIC
Temperature
Minimum: 78.0
K
Maximum: 80.0 K
Maximum: 80.0 K
Image Recording
[1]
Detector model:
GATAN K2 SUMMIT (4k x 4k)
Detector mode: SUPER-RESOLUTION
Dimensions: 7420 pixel x 7676 pixel
Frames per image: 3-16
Number of grids: 1
Number of real images: 1714
Average exposure time: 0.25 s
Average electron dose per image: 1.56 e/Å2
Detector mode: SUPER-RESOLUTION
Dimensions: 7420 pixel x 7676 pixel
Frames per image: 3-16
Number of grids: 1
Number of real images: 1714
Average exposure time: 0.25 s
Average electron dose per image: 1.56 e/Å2
Final
reconstruction
Resolution: 5.1
Å
(
BY AUTHOR)
Resolution method: FSC 0.143 CUT-OFF
Number of classed used: 2
Number of images used: 33813
Resolution method: FSC 0.143 CUT-OFF
Number of classed used: 2
Number of images used: 33813
⌯ Applied Symmetry
Point group:
C1
Software
[1]
| Name | Version | Details |
|---|---|---|
| RELION | 2 | - |
Startup model
[1]
⦨ Initial angle
assignment
⦩ Final angle assignment
Particle selection
[1]
| Selected | Ref. model | Method | Software | Details |
|---|---|---|---|---|
| 79414 | - | - | - | - |
Final 3D classification
Number of classes:
5
Software
[1]
| Name | Version | Details |
|---|---|---|
| RELION | 2 | - |
Format: CCP4
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: Yeast small subunit processome
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: Yeast small subunit processome
⬡ Geometry
| X | Y | Z | |
|---|---|---|---|
| Dimensions | 380 | 380 | 380 |
| Origin | 0 | 0 | 0 |
| Spacing | 380 | 380 | 380 |
| Voxel size | 1.325 Å | 1.325 Å | 1.325 Å |
Contour list
| Primary | Level | Source |
|---|---|---|
| True | 0.018 | AUTHOR |
