EMD-4584
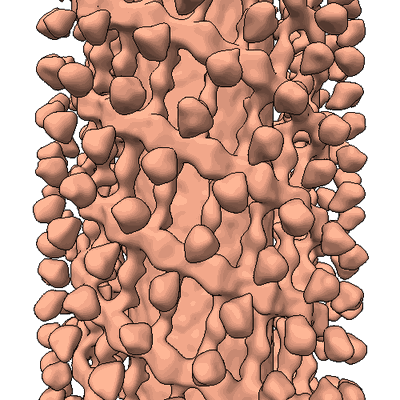
Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1
EMD-4584
Subtomogram averaging20.4 Å
 Deposition: 01/02/2019
Deposition: 01/02/2019Map released: 14/08/2019
Last modified: 25/11/2020
Concentration: 5
mg/mL
Details: Sample was prepared in the described buffer. Just before freezing 6 nm colloidal gold fiducial marker were added to the sample in a 1:1 ratio.
Details: Sample was prepared in the described buffer. Just before freezing 6 nm colloidal gold fiducial marker were added to the sample in a 1:1 ratio.
Buffer
pH: 7.4
Details: 20 mM HEPES, 200 mM NaCl, residual MgCl2, 9mM KCl.
Details: 20 mM HEPES, 200 mM NaCl, residual MgCl2, 9mM KCl.
Grid
Vitrification
Cryogen name: ETHANE
Chamber humidity: 70%
Chamber temperature: 277.15 K
Instrument: FEI VITROBOT MARK IV
Chamber humidity: 70%
Chamber temperature: 277.15 K
Instrument: FEI VITROBOT MARK IV
Microscope: FEI TITAN KRIOS
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 300 kV
C2 aperture diameter: 70.0 µm
Nominal CS: 2.7 mm
Nominal defocus: 2.0 µm - 4.0 µm
Nominal magnification: 53000.0
Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER
Cooling holder cryogen: NITROGEN
Alignment procedure: BASIC
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 300 kV
C2 aperture diameter: 70.0 µm
Nominal CS: 2.7 mm
Nominal defocus: 2.0 µm - 4.0 µm
Nominal magnification: 53000.0
Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER
Cooling holder cryogen: NITROGEN
Alignment procedure: BASIC
Specialist optics
Energy filter
Slit width:
20
eV
Image Recording
[1]
Detector model:
GATAN K2 SUMMIT (4k x 4k)
Detector mode: COUNTING
Average electron dose per image: 2.0 e/Å2
Detector mode: COUNTING
Average electron dose per image: 2.0 e/Å2
Final
reconstruction
Resolution: 20.4
Å
(
BY AUTHOR)
Resolution method: FSC 0.143 CUT-OFF
Resolution method: FSC 0.143 CUT-OFF
⌯ Applied Symmetry
Point group:
C1
Software
[1]
| Name | Version | Details |
|---|---|---|
| Dynamo | 1.1.266 | - |
⦩ Final angle assignment
Final 3D classification
Number of classes:
1
Avg. number of members per classes: 1677.0
Avg. number of members per classes: 1677.0
Software
[1]
| Name | Version | Details |
|---|---|---|
| Dynamo | 1.1.266 | - |
Extraction
Number of images used: 2214
Reference model: Global average of the particles rotated to the known initial orientations
Method: Geometry-assisted particle picking form tube surfaces
Reference model: Global average of the particles rotated to the known initial orientations
Method: Geometry-assisted particle picking form tube surfaces
Software
[1]
| Name | Version | Details |
|---|---|---|
| Dynamo | 1.1.266 | - |
Format: CCP4
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: None
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: None
⬡ Geometry
| X | Y | Z | |
|---|---|---|---|
| Dimensions | 280 | 280 | 280 |
| Origin | 0 | 0 | 0 |
| Spacing | 280 | 280 | 280 |
| Voxel size | 2.7 Å | 2.7 Å | 2.7 Å |
Contour list
| Primary | Level | Source |
|---|---|---|
| True | 1.0 | AUTHOR |
