EMD-2339
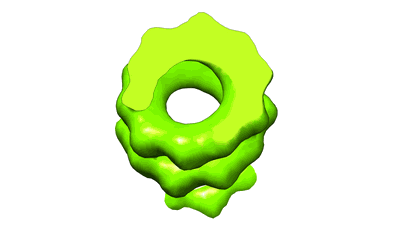
Variable internal flexibility characterizes the helical capsid formed by Agrobacterium VirE2 protein on single-stranded DNA.
EMD-2339
Helical reconstruction20.0 Å
 Deposition: 25/03/2013
Deposition: 25/03/2013Map released: 26/06/2013
Last modified: 17/07/2013
Concentration: 1
mg/mL
Details: Protein was mixed with single-stranded DNA
Details: Protein was mixed with single-stranded DNA
Buffer
pH: 8.0
Details: 50 mM Tris, 500 mM NaCl
Details: 50 mM Tris, 500 mM NaCl
Grid
Details: Quantifoil holey carbon
Vitrification
Microscope: FEI TECNAI F20
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 200 kV
Nominal CS: 2 mm
Nominal defocus: 1.0 µm - 3.2 µm
Nominal magnification: 50000.0
Specimen holder model: GATAN LIQUID NITROGEN
Alignment procedure: LEGACY (Astigmatism: Objective lens astigmatism was corrected at high-magnification (>100,000), Electron beam tilt params: )
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 200 kV
Nominal CS: 2 mm
Nominal defocus: 1.0 µm - 3.2 µm
Nominal magnification: 50000.0
Specimen holder model: GATAN LIQUID NITROGEN
Alignment procedure: LEGACY (Astigmatism: Objective lens astigmatism was corrected at high-magnification (>100,000), Electron beam tilt params: )
Image Recording
[1]
Detector category:
CCD
Detector model: GENERIC TVIPS
Average electron dose per image: 20 e/Å2
Details: Image data was collected as focal pairs.
Detector model: GENERIC TVIPS
Average electron dose per image: 20 e/Å2
Details: Image data was collected as focal pairs.
Details: Particles were picked and preselected using routines of Xmipp, and then reconstruction was carried out using IHRSR.
Final
reconstruction
Resolution: 20.0
Å
(
BY AUTHOR)
Resolution method: FSC 0.5 CUT-OFF
Details:
Resolution method: FSC 0.5 CUT-OFF
Details:
⌯ Applied Symmetry
ΔΖ:
14.67
Å
ΔΦ: 110.09°
ΔΦ: 110.09°
Software
[1]
| Name | Version | Details |
|---|---|---|
| Bsoft, EMAN, Xmipp, Spider, IHRSR | - | - |
CTF correction
Details:Phase-flipping
Format: CCP4
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: CryoEM reconstruction of the Agrobacterium T-complex
Details: ::::EMDATABANK.org::::EMD-2339::::
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: CryoEM reconstruction of the Agrobacterium T-complex
Details: ::::EMDATABANK.org::::EMD-2339::::
⬡ Geometry
| X | Y | Z | |
|---|---|---|---|
| Dimensions | 64 | 64 | 64 |
| Origin | 0 | 0 | 0 |
| Spacing | 64 | 64 | 64 |
| Voxel size | 4.32 Å | 4.32 Å | 4.32 Å |
Contour list
| Primary | Level | Source |
|---|---|---|
| True | 0.008 | AUTHOR |
