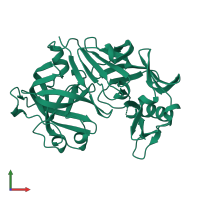
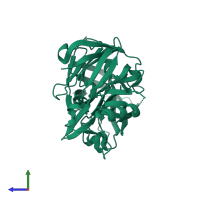
Function and Biology Details
Reaction catalysed:
Broad specificity similar to that of pepsin A. Clots milk by cleavage of a single 104-Ser-Phe-|-Met-Ala-107 bond in kappa-chain of casein.
Biochemical function:
Biological process:
Cellular component:
- not assigned
Sequence domains:
Structure domain:
Structure analysis Details
Assembly composition:
monomeric (preferred)
Assembly name:
Chymosin (preferred)
PDBe Complex ID:
PDB-CPX-133556 (preferred)
Entry contents:
1 distinct polypeptide molecule
Macromolecule:
Ligands and Environments
No bound ligands
No modified residues
Experiments and Validation Details
Spacegroup:
I222
Expression system: Not provided