Assemblies
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14648.66 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14877.28 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14984.76 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
15054.42 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14604.85 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14858.93 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14769.93 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14977.75 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14797.56 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14947.34 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14895.28 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Assembly Name:
MAP kinase-activated protein kinase 2
Multimeric state:
monomeric
Accessible surface area:
14967.4 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-155865
Macromolecules
Chains: A, B, C, D, E, F, G, H, I, J, K, L
Length: 318 amino acids
Theoretical weight: 36.75 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21
UniProt:
Pfam: Protein kinase domain
InterPro:
Length: 318 amino acids
Theoretical weight: 36.75 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21
UniProt:
- Canonical:
 P49137 (Residues: 47-364; Coverage: 80%)
P49137 (Residues: 47-364; Coverage: 80%)
Pfam: Protein kinase domain
InterPro:
- Protein kinase-like domain superfamily
- Protein kinase domain
- Protein kinase, ATP binding site
- Serine/threonine-protein kinase, active site
- MAP kinase activated protein kinase, C-terminal



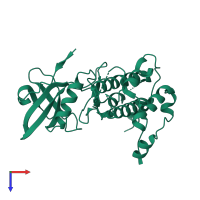
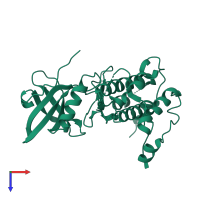
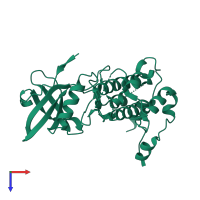
![The deposited structure of PDB entry <span class='highlight'>3r2b</span> coloured by chain, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>12 copies of <span class='highlight'>MAP kinase-activated protein kinase 2</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one</span>.</li></ul> PDB entry 3r2b coloured by chain, front view.](https://www.ebi.ac.uk/pdbe/static/entry/3r2b_deposited_chain_front_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>3r2b</span> coloured by chain, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>12 copies of <span class='highlight'>MAP kinase-activated protein kinase 2</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one</span>.</li></ul> PDB entry 3r2b coloured by chain, side view.](https://www.ebi.ac.uk/pdbe/static/entry/3r2b_deposited_chain_side_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>3r2b</span> coloured by chain, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>12 copies of <span class='highlight'>MAP kinase-activated protein kinase 2</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one</span>.</li></ul> PDB entry 3r2b coloured by chain, top view.](https://www.ebi.ac.uk/pdbe/static/entry/3r2b_deposited_chain_top_image-200x200.png)
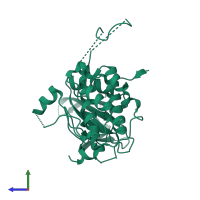
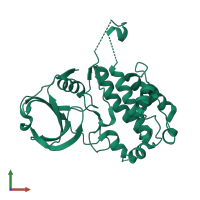
![Monomeric assembly 1 of PDB entry <span class='highlight'>3r2b</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>MAP kinase-activated protein kinase 2</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one</span>.</li></ul>} Monomeric assembly 1 of PDB entry 3r2b coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/3r2b_assembly_1_chemically_distinct_molecules_front_image-200x200.png)
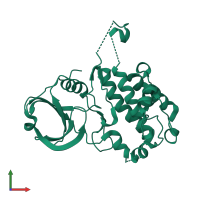
![Monomeric assembly 1 of PDB entry <span class='highlight'>3r2b</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>MAP kinase-activated protein kinase 2</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one</span>.</li></ul>} Monomeric assembly 1 of PDB entry 3r2b coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/3r2b_assembly_1_chemically_distinct_molecules_side_image-200x200.png)
![Monomeric assembly 1 of PDB entry <span class='highlight'>3r2b</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>MAP kinase-activated protein kinase 2</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one</span>.</li></ul>} Monomeric assembly 1 of PDB entry 3r2b coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/3r2b_assembly_1_chemically_distinct_molecules_top_image-200x200.png)
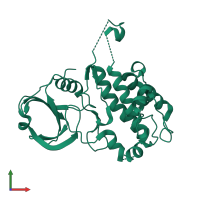








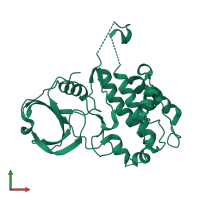
![Monomeric assembly 2 of PDB entry <span class='highlight'>3r2b</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>MAP kinase-activated protein kinase 2</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one</span>.</li></ul>} Monomeric assembly 2 of PDB entry 3r2b coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/3r2b_assembly_2_chemically_distinct_molecules_front_image-200x200.png)
![Monomeric assembly 2 of PDB entry <span class='highlight'>3r2b</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>MAP kinase-activated protein kinase 2</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one</span>.</li></ul>} Monomeric assembly 2 of PDB entry 3r2b coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/3r2b_assembly_2_chemically_distinct_molecules_side_image-200x200.png)
![Monomeric assembly 2 of PDB entry <span class='highlight'>3r2b</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>MAP kinase-activated protein kinase 2</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one</span>.</li></ul>} Monomeric assembly 2 of PDB entry 3r2b coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/3r2b_assembly_2_chemically_distinct_molecules_top_image-200x200.png)