Assemblies
Multimeric state:
hetero hexamer
Accessible surface area:
69640.56 Å2
Buried surface area:
36270.6 Å2
Dissociation area:
3,331.56
Å2
Dissociation energy (ΔGdiss):
57.03
kcal/mol
Dissociation entropy (TΔSdiss):
16.69
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-169747
Multimeric state:
hetero hexamer
Accessible surface area:
69801.21 Å2
Buried surface area:
36183.12 Å2
Dissociation area:
3,320.71
Å2
Dissociation energy (ΔGdiss):
58.16
kcal/mol
Dissociation entropy (TΔSdiss):
16.68
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-169747
Multimeric state:
hetero hexamer
Accessible surface area:
69631.82 Å2
Buried surface area:
36345.68 Å2
Dissociation area:
3,343.14
Å2
Dissociation energy (ΔGdiss):
58.13
kcal/mol
Dissociation entropy (TΔSdiss):
16.69
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-169747
Macromolecules
Chains: A, D, G, J, M, P
Length: 428 amino acids
Theoretical weight: 48.44 KDa
Source organism: Cereibacter sphaeroides
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
Length: 428 amino acids
Theoretical weight: 48.44 KDa
Source organism: Cereibacter sphaeroides
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q02761 (Residues: 3-430; Coverage: 96%)
Q02761 (Residues: 3-430; Coverage: 96%)
Pfam:
InterPro:
- Cytochrome b/b6-like domain superfamily
- Cytochrome b
- Cytochrome b/b6, N-terminal domain
- Di-haem cytochrome, transmembrane
- Cytochrome b/b6, C-terminal
- Cytochrome b/b6, C-terminal domain superfamily
Chains: B, E, H, K, N, Q
Length: 256 amino acids
Theoretical weight: 27.79 KDa
Source organism: Cereibacter sphaeroides
Expression system: Escherichia coli
UniProt:
Pfam: Cytochrome C1 family
InterPro:
CATH:
Length: 256 amino acids
Theoretical weight: 27.79 KDa
Source organism: Cereibacter sphaeroides
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q02760 (Residues: 23-278; Coverage: 97%)
Q02760 (Residues: 23-278; Coverage: 97%)
Pfam: Cytochrome C1 family
InterPro:
CATH:
Chains: C, F, I, L, O, R
Length: 179 amino acids
Theoretical weight: 19.06 KDa
Source organism: Cereibacter sphaeroides
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
Length: 179 amino acids
Theoretical weight: 19.06 KDa
Source organism: Cereibacter sphaeroides
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q02762 (Residues: 9-187; Coverage: 96%)
Q02762 (Residues: 9-187; Coverage: 96%)
Pfam:
InterPro:
- Ubiquinol-cytochrome c reductase, iron-sulphur subunit
- Rieske iron-sulphur protein
- Ubiquitinol-cytochrome C reductase, Fe-S subunit, TAT signal
- Twin-arginine translocation pathway, signal sequence, bacterial/archaeal
- Rieske [2Fe-2S] iron-sulphur domain superfamily
- Rieske [2Fe-2S] iron-sulphur domain
- Rieske iron-sulphur protein, C-terminal



![The deposited structure of PDB entry <span class='highlight'>2qjk</span> coloured by chain, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>6 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>18 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul> PDB entry 2qjk coloured by chain, front view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_deposited_chain_front_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>2qjk</span> coloured by chain, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>6 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>18 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul> PDB entry 2qjk coloured by chain, side view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_deposited_chain_side_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>2qjk</span> coloured by chain, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>6 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>18 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul> PDB entry 2qjk coloured by chain, top view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_deposited_chain_top_image-200x200.png)
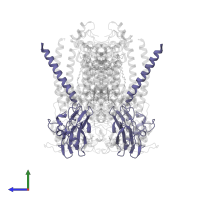
![Hetero hexameric assembly 1 of PDB entry <span class='highlight'>2qjk</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul>} Hetero hexameric assembly 1 of PDB entry 2qjk coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_assembly_1_chemically_distinct_molecules_front_image-200x200.png)
![Hetero hexameric assembly 1 of PDB entry <span class='highlight'>2qjk</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul>} Hetero hexameric assembly 1 of PDB entry 2qjk coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_assembly_1_chemically_distinct_molecules_side_image-200x200.png)
![Hetero hexameric assembly 1 of PDB entry <span class='highlight'>2qjk</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul>} Hetero hexameric assembly 1 of PDB entry 2qjk coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_assembly_1_chemically_distinct_molecules_top_image-200x200.png)
![Hetero hexameric assembly 2 of PDB entry <span class='highlight'>2qjk</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul>} Hetero hexameric assembly 2 of PDB entry 2qjk coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_assembly_2_chemically_distinct_molecules_front_image-200x200.png)
![Hetero hexameric assembly 2 of PDB entry <span class='highlight'>2qjk</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul>} Hetero hexameric assembly 2 of PDB entry 2qjk coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_assembly_2_chemically_distinct_molecules_side_image-200x200.png)
![Hetero hexameric assembly 2 of PDB entry <span class='highlight'>2qjk</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul>} Hetero hexameric assembly 2 of PDB entry 2qjk coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_assembly_2_chemically_distinct_molecules_top_image-200x200.png)
![Hetero hexameric assembly 3 of PDB entry <span class='highlight'>2qjk</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul>} Hetero hexameric assembly 3 of PDB entry 2qjk coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_assembly_3_chemically_distinct_molecules_front_image-200x200.png)
![Hetero hexameric assembly 3 of PDB entry <span class='highlight'>2qjk</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul>} Hetero hexameric assembly 3 of PDB entry 2qjk coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_assembly_3_chemically_distinct_molecules_side_image-200x200.png)
![Hetero hexameric assembly 3 of PDB entry <span class='highlight'>2qjk</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome b</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Cytochrome c1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Ubiquinol-cytochrome c reductase iron-sulfur subunit</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>2-O-octyl-beta-D-glucopyranose</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>PROTOPORPHYRIN IX CONTAINING FE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STIGMATELLIN A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>(2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>STRONTIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>FE2/S2 (INORGANIC) CLUSTER</span>.</li></ul>} Hetero hexameric assembly 3 of PDB entry 2qjk coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/2qjk_assembly_3_chemically_distinct_molecules_top_image-200x200.png)