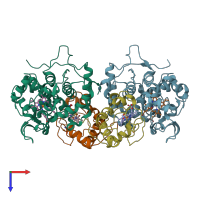
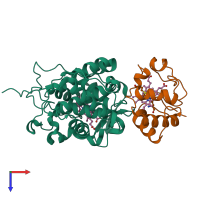
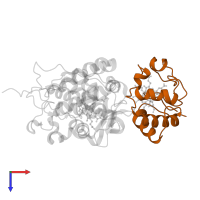
Assemblies
Multimeric state:
hetero dimer
Accessible surface area:
17211.21 Å2
Buried surface area:
3861.08 Å2
Dissociation area:
600.51
Å2
Dissociation energy (ΔGdiss):
-7.04
kcal/mol
Dissociation entropy (TΔSdiss):
11.62
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-132231
Multimeric state:
hetero dimer
Accessible surface area:
17267.19 Å2
Buried surface area:
3799.2 Å2
Dissociation area:
568.09
Å2
Dissociation energy (ΔGdiss):
-6.53
kcal/mol
Dissociation entropy (TΔSdiss):
11.6
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-132231
Macromolecules
Chains: A, C
Length: 296 amino acids
Theoretical weight: 33.77 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Not provided
UniProt:
Pfam: Peroxidase
InterPro:
SCOP: CCP-like
Length: 296 amino acids
Theoretical weight: 33.77 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Not provided
UniProt:
- Canonical:
 P00431 (Residues: 68-361; Coverage: 81%)
P00431 (Residues: 68-361; Coverage: 81%)
Pfam: Peroxidase
InterPro:
- Haem peroxidase superfamily
- Heme-binding peroxidase Ccp1-like
- Haem peroxidase
- Class I peroxidase
- Peroxidase, active site
- Peroxidases heam-ligand binding site
SCOP: CCP-like
Chains: B, D
Length: 108 amino acids
Theoretical weight: 12.07 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Not provided
UniProt:
Pfam: Cytochrome c
InterPro:
CATH: Cytochrome c-like domain
SCOP: monodomain cytochrome c
Length: 108 amino acids
Theoretical weight: 12.07 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Not provided
UniProt:
- Canonical:
 P00044 (Residues: 2-109; Coverage: 99%)
P00044 (Residues: 2-109; Coverage: 99%)
Pfam: Cytochrome c
InterPro:
CATH: Cytochrome c-like domain
SCOP: monodomain cytochrome c