Assemblies
Assembly Name:
Nuclear receptor coactivator 1 and Vitamin D3 receptor A
Multimeric state:
hetero tetramer
Accessible surface area:
21759.03 Å2
Buried surface area:
6215.29 Å2
Dissociation area:
2,100.91
Å2
Dissociation energy (ΔGdiss):
9.77
kcal/mol
Dissociation entropy (TΔSdiss):
27.86
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-172403
Macromolecules
Chain: A
Length: 302 amino acids
Theoretical weight: 34.06 KDa
Source organism: Danio rerio
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Ligand-binding domain of nuclear hormone receptor
InterPro:
Length: 302 amino acids
Theoretical weight: 34.06 KDa
Source organism: Danio rerio
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q9PTN2 (Residues: 156-453; Coverage: 66%)
Q9PTN2 (Residues: 156-453; Coverage: 66%)
Pfam: Ligand-binding domain of nuclear hormone receptor
InterPro:
- Nuclear hormone receptor-like domain superfamily
- Nuclear hormone receptor, ligand-binding domain
- Vitamin D receptor
- Nuclear hormone receptor
Chain: B
Length: 15 amino acids
Theoretical weight: 1.78 KDa
Source organism: Homo sapiens
Expression system: Not provided
UniProt:
InterPro:
Length: 15 amino acids
Theoretical weight: 1.78 KDa
Source organism: Homo sapiens
Expression system: Not provided
UniProt:
- Canonical:
 Q15788 (Residues: 686-700; Coverage: 1%)
Q15788 (Residues: 686-700; Coverage: 1%)
InterPro:



![The deposited structure of PDB entry <span class='highlight'>2hbh</span> coloured by chain, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Vitamin D3 receptor A</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Nuclear receptor coactivator 1</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>1,3-CYCLOHEXANEDIOL, 4-METHYLENE-5-[(2E)-[(1S,3AS,7AS)-OCTAHYDRO-1-(5-HYDROXY-5-METHYL-1,3-HEXADIYNYL)-7A-METHYL-4H-INDEN-4-YLIDENE]ETHYLIDENE]-, (1R,3S,5Z)</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 2hbh coloured by chain, front view.](https://www.ebi.ac.uk/pdbe/static/entry/2hbh_deposited_chain_front_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>2hbh</span> coloured by chain, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Vitamin D3 receptor A</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Nuclear receptor coactivator 1</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>1,3-CYCLOHEXANEDIOL, 4-METHYLENE-5-[(2E)-[(1S,3AS,7AS)-OCTAHYDRO-1-(5-HYDROXY-5-METHYL-1,3-HEXADIYNYL)-7A-METHYL-4H-INDEN-4-YLIDENE]ETHYLIDENE]-, (1R,3S,5Z)</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 2hbh coloured by chain, side view.](https://www.ebi.ac.uk/pdbe/static/entry/2hbh_deposited_chain_side_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>2hbh</span> coloured by chain, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Vitamin D3 receptor A</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Nuclear receptor coactivator 1</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>1,3-CYCLOHEXANEDIOL, 4-METHYLENE-5-[(2E)-[(1S,3AS,7AS)-OCTAHYDRO-1-(5-HYDROXY-5-METHYL-1,3-HEXADIYNYL)-7A-METHYL-4H-INDEN-4-YLIDENE]ETHYLIDENE]-, (1R,3S,5Z)</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 2hbh coloured by chain, top view.](https://www.ebi.ac.uk/pdbe/static/entry/2hbh_deposited_chain_top_image-200x200.png)
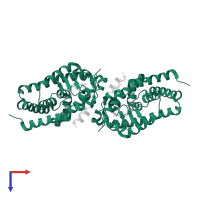
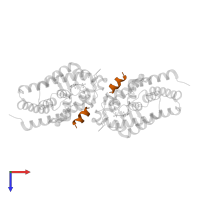
![Hetero tetrameric assembly 1 of PDB entry <span class='highlight'>2hbh</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Vitamin D3 receptor A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Nuclear receptor coactivator 1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>1,3-CYCLOHEXANEDIOL, 4-METHYLENE-5-[(2E)-[(1S,3AS,7AS)-OCTAHYDRO-1-(5-HYDROXY-5-METHYL-1,3-HEXADIYNYL)-7A-METHYL-4H-INDEN-4-YLIDENE]ETHYLIDENE]-, (1R,3S,5Z)</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>water</span>.</li></ul>} Hetero tetrameric assembly 1 of PDB entry 2hbh coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/2hbh_assembly_1_chemically_distinct_molecules_front_image-200x200.png)
![Hetero tetrameric assembly 1 of PDB entry <span class='highlight'>2hbh</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Vitamin D3 receptor A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Nuclear receptor coactivator 1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>1,3-CYCLOHEXANEDIOL, 4-METHYLENE-5-[(2E)-[(1S,3AS,7AS)-OCTAHYDRO-1-(5-HYDROXY-5-METHYL-1,3-HEXADIYNYL)-7A-METHYL-4H-INDEN-4-YLIDENE]ETHYLIDENE]-, (1R,3S,5Z)</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>water</span>.</li></ul>} Hetero tetrameric assembly 1 of PDB entry 2hbh coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/2hbh_assembly_1_chemically_distinct_molecules_side_image-200x200.png)
![Hetero tetrameric assembly 1 of PDB entry <span class='highlight'>2hbh</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Vitamin D3 receptor A</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Nuclear receptor coactivator 1</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>1,3-CYCLOHEXANEDIOL, 4-METHYLENE-5-[(2E)-[(1S,3AS,7AS)-OCTAHYDRO-1-(5-HYDROXY-5-METHYL-1,3-HEXADIYNYL)-7A-METHYL-4H-INDEN-4-YLIDENE]ETHYLIDENE]-, (1R,3S,5Z)</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>water</span>.</li></ul>} Hetero tetrameric assembly 1 of PDB entry 2hbh coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/2hbh_assembly_1_chemically_distinct_molecules_top_image-200x200.png)