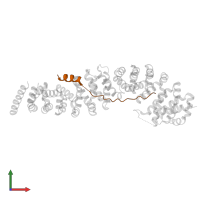
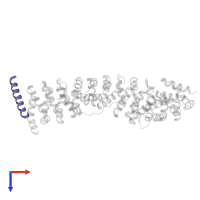
Assemblies
Multimeric state:
hetero trimer
Accessible surface area:
23963.08 Å2
Buried surface area:
4613.51 Å2
Dissociation area:
604.83
Å2
Dissociation energy (ΔGdiss):
7.39
kcal/mol
Dissociation entropy (TΔSdiss):
8.99
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-126328
Multimeric state:
hetero trimer
Accessible surface area:
23705.99 Å2
Buried surface area:
4024.84 Å2
Dissociation area:
396.74
Å2
Dissociation energy (ΔGdiss):
1.29
kcal/mol
Dissociation entropy (TΔSdiss):
8.29
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-126328
Macromolecules
Chains: A, D
Length: 550 amino acids
Theoretical weight: 60.24 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Armadillo/beta-catenin-like repeat
InterPro:
CATH: Leucine-rich Repeat Variant
SCOP: Armadillo repeat
Length: 550 amino acids
Theoretical weight: 60.24 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P35222 (Residues: 138-686; Coverage: 70%)
P35222 (Residues: 138-686; Coverage: 70%)
Pfam: Armadillo/beta-catenin-like repeat
InterPro:
CATH: Leucine-rich Repeat Variant
SCOP: Armadillo repeat
Chains: B, E
Length: 53 amino acids
Theoretical weight: 5.63 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
InterPro:
Length: 53 amino acids
Theoretical weight: 5.63 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q9NQB0 (Residues: 1-53; Coverage: 9%)
Q9NQB0 (Residues: 1-53; Coverage: 9%)
InterPro:
Chains: C, F
Length: 46 amino acids
Theoretical weight: 5.29 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: B-cell lymphoma 9 protein
InterPro:
Length: 46 amino acids
Theoretical weight: 5.29 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 O00512 (Residues: 347-392; Coverage: 3%)
O00512 (Residues: 347-392; Coverage: 3%)
Pfam: B-cell lymphoma 9 protein
InterPro: