Assemblies
Assembly Name:
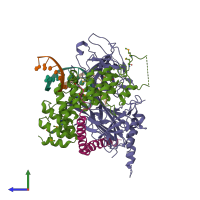
CSL-Notch-Mastermind transcription factor complex and DNA
Multimeric state:
hetero pentamer
Accessible surface area:
40040.47 Å2
Buried surface area:
12825.94 Å2
Dissociation area:
2,244.89
Å2
Dissociation energy (ΔGdiss):
-1.49
kcal/mol
Dissociation entropy (TΔSdiss):
12.6
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-115273
Macromolecules
Chain: A
Length: 477 amino acids
Theoretical weight: 53.76 KDa
Source organism: Caenorhabditis elegans
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
SCOP:
Length: 477 amino acids
Theoretical weight: 53.76 KDa
Source organism: Caenorhabditis elegans
Expression system: Escherichia coli
UniProt:
- Canonical:
 V6CLJ5 (Residues: 309-780; Coverage: 60%)
V6CLJ5 (Residues: 309-780; Coverage: 60%)
Pfam:
InterPro:
- p53-like transcription factor, DNA-binding domain superfamily
- RBP-J/Cbf11, DNA binding domain superfamily
- Suppressor of hairless-like
- RBP-J/Cbf11/Cbf12, DNA binding
- Beta-trefoil domain superfamily
- Beta-trefoil DNA-binding domain
- Immunoglobulin E-set
- RBP-Jkappa, IPT domain
- Immunoglobulin-like fold
SCOP:
Chain: D
Length: 85 amino acids
Theoretical weight: 9.76 KDa
Source organism: Caenorhabditis elegans
Expression system: Escherichia coli
UniProt:
InterPro: Transcription activator LAG-3
CATH: Single alpha-helices involved in coiled-coils or other helix-helix interfaces
SCOP: Lag-3 N-terminal region
Length: 85 amino acids
Theoretical weight: 9.76 KDa
Source organism: Caenorhabditis elegans
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q09260 (Residues: 49-132; Coverage: 17%)
Q09260 (Residues: 49-132; Coverage: 17%)
InterPro: Transcription activator LAG-3
CATH: Single alpha-helices involved in coiled-coils or other helix-helix interfaces
SCOP: Lag-3 N-terminal region
Chain: E
Length: 373 amino acids
Theoretical weight: 42.46 KDa
Source organism: Caenorhabditis elegans
Expression system: Escherichia coli
UniProt:
Pfam: Ankyrin repeat
InterPro:
CATH: Ankyrin repeat-containing domain
SCOP: Ankyrin repeat
Length: 373 amino acids
Theoretical weight: 42.46 KDa
Source organism: Caenorhabditis elegans
Expression system: Escherichia coli
UniProt:
- Canonical:
 P14585 (Residues: 931-1301; Coverage: 26%)
P14585 (Residues: 931-1301; Coverage: 26%)
Pfam: Ankyrin repeat
InterPro:
CATH: Ankyrin repeat-containing domain
SCOP: Ankyrin repeat
Name:
5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'
Representative chains: B
Length: 15 nucleotides
Theoretical weight: 4.67 KDa
Representative chains: B
Length: 15 nucleotides
Theoretical weight: 4.67 KDa
Name:
5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'
Representative chains: C
Length: 15 nucleotides
Theoretical weight: 4.5 KDa
Representative chains: C
Length: 15 nucleotides
Theoretical weight: 4.5 KDa