Assemblies
Assembly Name:
Peptidyl-prolyl cis-trans isomerase A
Multimeric state:
monomeric
Accessible surface area:
7869.94 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-142738
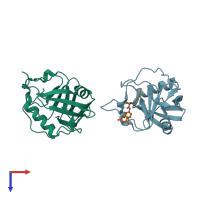
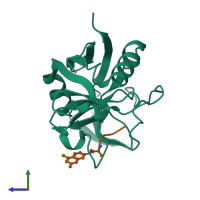
Assembly Name:
Peptidyl-prolyl cis-trans isomerase A and peptide
Multimeric state:
hetero dimer
Accessible surface area:
7308.6 Å2
Buried surface area:
1166.94 Å2
Dissociation area:
474.96
Å2
Dissociation energy (ΔGdiss):
-0.69
kcal/mol
Dissociation entropy (TΔSdiss):
5.63
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-142740
Macromolecules
Chains: A, B
Length: 166 amino acids
Theoretical weight: 18.07 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
Pfam: Cyclophilin type peptidyl-prolyl cis-trans isomerase/CLD
InterPro:
SCOP: Cyclophilin (peptidylprolyl isomerase)
Length: 166 amino acids
Theoretical weight: 18.07 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
- Canonical:
 P0AFL3 (Residues: 25-190; Coverage: 100%)
P0AFL3 (Residues: 25-190; Coverage: 100%)
Pfam: Cyclophilin type peptidyl-prolyl cis-trans isomerase/CLD
InterPro:
- Cyclophilin-like domain superfamily
- Cyclophilin-type peptidyl-prolyl cis-trans isomerase, E. coli cyclophilin A-like
- Cyclophilin-type peptidyl-prolyl cis-trans isomerase
- Cyclophilin-type peptidyl-prolyl cis-trans isomerase domain
- Cyclophilin-type peptidyl-prolyl cis-trans isomerase, conserved site
SCOP: Cyclophilin (peptidylprolyl isomerase)
Chain: C
Length: 6 amino acids
Theoretical weight: 512 Da
Source organism: Escherichia coli
Expression system: Not provided
Length: 6 amino acids
Theoretical weight: 512 Da
Source organism: Escherichia coli
Expression system: Not provided