Assemblies
Assembly Name:
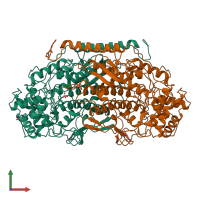
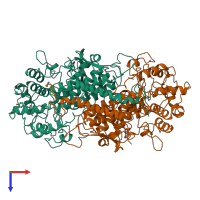
Vanadium-dependent bromoperoxidase
Multimeric state:
hetero dimer
Accessible surface area:
34596.16 Å2
Buried surface area:
11102.43 Å2
Dissociation area:
5,332.17
Å2
Dissociation energy (ΔGdiss):
72.36
kcal/mol
Dissociation entropy (TΔSdiss):
15.93
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-223924
Macromolecules
Chain: A
Length: 556 amino acids
Theoretical weight: 60.66 KDa
Source organism: Ascophyllum nodosum
UniProt:
SCOP: Haloperoxidase (bromoperoxidase)
Length: 556 amino acids
Theoretical weight: 60.66 KDa
Source organism: Ascophyllum nodosum
UniProt:
- Canonical:
 P81701 (Residues: 1-556; Coverage: 100%)
P81701 (Residues: 1-556; Coverage: 100%)
- Bromoperoxidase/chloroperoxidase C-terminal
- Phosphatidic acid phosphatase type 2/haloperoxidase superfamily
SCOP: Haloperoxidase (bromoperoxidase)
Chain: B
Length: 556 amino acids
Theoretical weight: 60.54 KDa
Source organism: Ascophyllum nodosum
UniProt:
SCOP: Haloperoxidase (bromoperoxidase)
Length: 556 amino acids
Theoretical weight: 60.54 KDa
Source organism: Ascophyllum nodosum
UniProt:
- Canonical:
 P81701 (Residues: 1-556; Coverage: 100%)
P81701 (Residues: 1-556; Coverage: 100%)
- Bromoperoxidase/chloroperoxidase C-terminal
- Phosphatidic acid phosphatase type 2/haloperoxidase superfamily
SCOP: Haloperoxidase (bromoperoxidase)