Assemblies
Assembly Name:
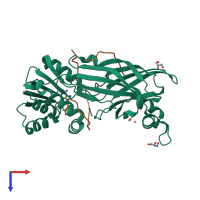
Protein arginine N-methyltransferase 1 and peptide
Multimeric state:
hetero pentamer
Accessible surface area:
14707.07 Å2
Buried surface area:
746.33 Å2
Dissociation area:
162.18
Å2
Dissociation energy (ΔGdiss):
-6.1
kcal/mol
Dissociation entropy (TΔSdiss):
0.72
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-162354
Assembly Name:
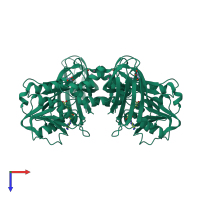
Protein arginine N-methyltransferase 1
Multimeric state:
homo dimer
Accessible surface area:
26148.04 Å2
Buried surface area:
4758.77 Å2
Dissociation area:
1,633.05
Å2
Dissociation energy (ΔGdiss):
21.4
kcal/mol
Dissociation entropy (TΔSdiss):
13.95
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-178989
Macromolecules
Chain: A
Length: 340 amino acids
Theoretical weight: 39.27 KDa
Source organism: Rattus norvegicus
Expression system: Escherichia coli
UniProt:
Pfam: Methyltransferase domain
InterPro:
SCOP: Arginine methyltransferase
Length: 340 amino acids
Theoretical weight: 39.27 KDa
Source organism: Rattus norvegicus
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q63009 (Residues: 14-353; Coverage: 96%)
Q63009 (Residues: 14-353; Coverage: 96%)
Pfam: Methyltransferase domain
InterPro:
- Protein arginine N-methyltransferase
- S-adenosyl-L-methionine-dependent methyltransferase superfamily
- Methyltransferase domain 25
SCOP: Arginine methyltransferase
Chains: B, C, D, E
Length: 19 amino acids
Theoretical weight: 1.67 KDa
Source organism: Rattus norvegicus
Expression system: Not provided
Length: 19 amino acids
Theoretical weight: 1.67 KDa
Source organism: Rattus norvegicus
Expression system: Not provided