Assemblies
Assembly Name:
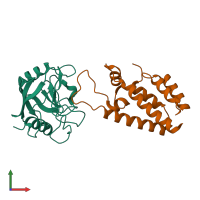
Peptidyl-prolyl cis-trans isomerase A and Gag Polyprotein
Multimeric state:
hetero dimer
Accessible surface area:
14626.54 Å2
Buried surface area:
794.64 Å2
Dissociation area:
397.32
Å2
Dissociation energy (ΔGdiss):
-3.53
kcal/mol
Dissociation entropy (TΔSdiss):
11.2
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-158799
Assembly Name:
Peptidyl-prolyl cis-trans isomerase A and Gag Polyprotein
Multimeric state:
hetero dimer
Accessible surface area:
14736.83 Å2
Buried surface area:
1091.48 Å2
Dissociation area:
545.74
Å2
Dissociation energy (ΔGdiss):
-4.96
kcal/mol
Dissociation entropy (TΔSdiss):
11.33
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-158799
Macromolecules
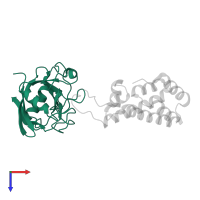
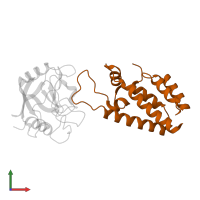
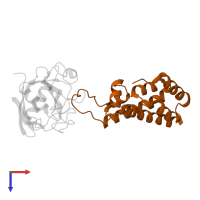
Chains: A, B
Length: 164 amino acids
Theoretical weight: 17.91 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Cyclophilin type peptidyl-prolyl cis-trans isomerase/CLD
InterPro:
SCOP: Cyclophilin (peptidylprolyl isomerase)
Length: 164 amino acids
Theoretical weight: 17.91 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P62937 (Residues: 1-164; Coverage: 99%)
P62937 (Residues: 1-164; Coverage: 99%)
Pfam: Cyclophilin type peptidyl-prolyl cis-trans isomerase/CLD
InterPro:
- Cyclophilin-type peptidyl-prolyl cis-trans isomerase
- Cyclophilin-like domain superfamily
- Cyclophilin-type peptidyl-prolyl cis-trans isomerase domain
- Cyclophilin-type peptidyl-prolyl cis-trans isomerase, conserved site
SCOP: Cyclophilin (peptidylprolyl isomerase)
Chains: C, D
Length: 146 amino acids
Theoretical weight: 16.14 KDa
Source organism: Human immunodeficiency virus 1
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: gag protein p24 N-terminal domain
InterPro:
CATH: Human Immunodeficiency Virus Type 1 Capsid Protein
SCOP: Retrovirus capsid protein, N-terminal core domain
Length: 146 amino acids
Theoretical weight: 16.14 KDa
Source organism: Human immunodeficiency virus 1
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q72497 (Residues: 133-278; Coverage: 29%)
Q72497 (Residues: 133-278; Coverage: 29%)
Pfam: gag protein p24 N-terminal domain
InterPro:
CATH: Human Immunodeficiency Virus Type 1 Capsid Protein
SCOP: Retrovirus capsid protein, N-terminal core domain