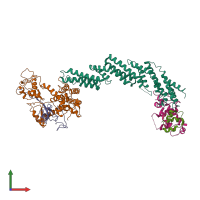
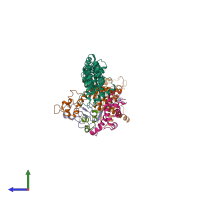
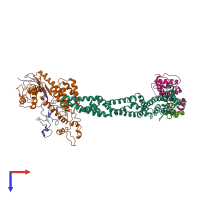
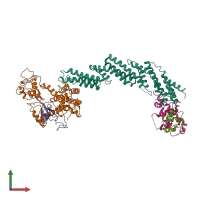
Assemblies
Assembly Name:
SCF E3 ubiquitin ligase complex, SKP2 variant
Multimeric state:
hetero pentamer
Accessible surface area:
51511.11 Å2
Buried surface area:
9967.28 Å2
Dissociation area:
938.19
Å2
Dissociation energy (ΔGdiss):
2.2
kcal/mol
Dissociation entropy (TΔSdiss):
14.33
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-158744
Multimeric state:
hetero decamer
Accessible surface area:
99810.9 Å2
Buried surface area:
23157.85 Å2
Dissociation area:
1,609.78
Å2
Dissociation energy (ΔGdiss):
5.64
kcal/mol
Dissociation entropy (TΔSdiss):
17.13
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-158752
Macromolecules
Chain: A
Length: 396 amino acids
Theoretical weight: 45.75 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Cullin family
InterPro:
CATH: Cullin Repeats
SCOP: Cullin repeat
Length: 396 amino acids
Theoretical weight: 45.75 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q13616 (Residues: 15-410; Coverage: 51%)
Q13616 (Residues: 15-410; Coverage: 51%)
Pfam: Cullin family
InterPro:
CATH: Cullin Repeats
SCOP: Cullin repeat
Chain: B
Length: 366 amino acids
Theoretical weight: 42.51 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
Length: 366 amino acids
Theoretical weight: 42.51 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q13616 (Residues: 411-776; Coverage: 47%)
Q13616 (Residues: 411-776; Coverage: 47%)
Pfam:
InterPro:
- Cullin
- Cullin, N-terminal
- Cullin homology domain superfamily
- Cullin homology domain
- Winged helix-like DNA-binding domain superfamily
- Winged helix DNA-binding domain superfamily
- Cullin protein, neddylation domain
- Cullin, conserved site
- Cullin Repeats
- Cullin; Chain C, Domain 2
- Winged helix-like DNA-binding domain superfamily/Winged helix DNA-binding domain
Chain: C
Length: 90 amino acids
Theoretical weight: 10.66 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: RING-H2 zinc finger domain
InterPro:
CATH: Zinc/RING finger domain, C3HC4 (zinc finger)
SCOP: RING finger domain, C3HC4
Length: 90 amino acids
Theoretical weight: 10.66 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P62877 (Residues: 19-108; Coverage: 83%)
P62877 (Residues: 19-108; Coverage: 83%)
Pfam: RING-H2 zinc finger domain
InterPro:
CATH: Zinc/RING finger domain, C3HC4 (zinc finger)
SCOP: RING finger domain, C3HC4
Chain: D
Length: 133 amino acids
Theoretical weight: 15.03 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
SCOP:
Length: 133 amino acids
Theoretical weight: 15.03 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P63208 (Residues: 2-140; Coverage: 82%)
P63208 (Residues: 2-140; Coverage: 82%)
Pfam:
InterPro:
- S-phase kinase-associated protein 1-like
- SKP1/BTB/POZ domain superfamily
- SKP1 component, POZ domain
- S-phase kinase-associated protein 1
- SKP1-like, dimerisation domain superfamily
- SKP1 component, dimerisation
SCOP:
Chain: E
Length: 41 amino acids
Theoretical weight: 4.76 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: F-box-like
InterPro:
CATH: Monooxygenase
SCOP: F-box domain
Length: 41 amino acids
Theoretical weight: 4.76 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q13309 (Residues: 97-137; Coverage: 10%)
Q13309 (Residues: 97-137; Coverage: 10%)
Pfam: F-box-like
InterPro:
CATH: Monooxygenase
SCOP: F-box domain