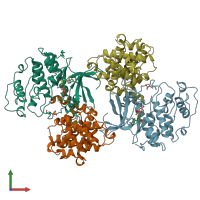
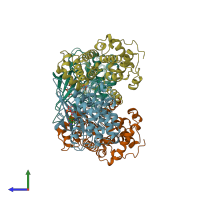
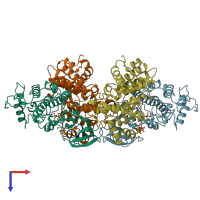
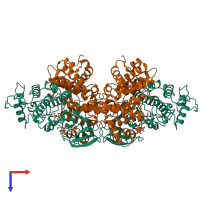
Assemblies
Assembly Name:
Cyclin-A2 and Cyclin-dependent kinase 2
Multimeric state:
hetero tetramer
Accessible surface area:
42632.34 Å2
Buried surface area:
10388.43 Å2
Dissociation area:
952.2
Å2
Dissociation energy (ΔGdiss):
-7.03
kcal/mol
Dissociation entropy (TΔSdiss):
14.99
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-148908
Assembly Name:
Cyclin A2-CDK2 complex
Multimeric state:
hetero dimer
Accessible surface area:
22212.88 Å2
Buried surface area:
4286.15 Å2
Dissociation area:
1,643.11
Å2
Dissociation energy (ΔGdiss):
14.17
kcal/mol
Dissociation entropy (TΔSdiss):
13.18
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-148886
Assembly Name:
Cyclin A2-CDK2 complex
Multimeric state:
hetero dimer
Accessible surface area:
22323.86 Å2
Buried surface area:
4197.88 Å2
Dissociation area:
1,602.07
Å2
Dissociation energy (ΔGdiss):
13.97
kcal/mol
Dissociation entropy (TΔSdiss):
13.17
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-148886
Macromolecules
Chains: A, C
Length: 298 amino acids
Theoretical weight: 34.06 KDa
Source organism: Homo sapiens
Expression system: Spodoptera frugiperda
UniProt:
Pfam: Protein kinase domain
InterPro:
SCOP: Protein kinases, catalytic subunit
Length: 298 amino acids
Theoretical weight: 34.06 KDa
Source organism: Homo sapiens
Expression system: Spodoptera frugiperda
UniProt:
- Canonical:
 P24941 (Residues: 1-298; Coverage: 100%)
P24941 (Residues: 1-298; Coverage: 100%)
Pfam: Protein kinase domain
InterPro:
- Protein kinase-like domain superfamily
- Protein kinase domain
- Protein kinase, ATP binding site
- Serine/threonine-protein kinase, active site
SCOP: Protein kinases, catalytic subunit
Chains: B, D
Length: 258 amino acids
Theoretical weight: 29.62 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
SCOP: Cyclin
Length: 258 amino acids
Theoretical weight: 29.62 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P20248 (Residues: 175-432; Coverage: 60%)
P20248 (Residues: 175-432; Coverage: 60%)
Pfam:
InterPro:
- Cyclin A/B-like
- Cyclin-like superfamily
- Cyclin
- Cyclin, N-terminal
- Cyclin-like domain
- Cyclin, C-terminal domain
SCOP: Cyclin