Assemblies
Assembly Name:
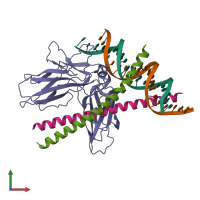
AP-1 transcription factor complex FOS-JUN-NFATC2 and DNA
Multimeric state:
hetero pentamer
Accessible surface area:
26405.34 Å2
Buried surface area:
9519.07 Å2
Dissociation area:
2,924.85
Å2
Dissociation energy (ΔGdiss):
17.97
kcal/mol
Dissociation entropy (TΔSdiss):
25.08
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-113378
Macromolecules
Chain: N
Length: 301 amino acids
Theoretical weight: 34.27 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam:
InterPro:
SCOP:
Length: 301 amino acids
Theoretical weight: 34.27 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q13469 (Residues: 392-678; Coverage: 31%)
Q13469 (Residues: 392-678; Coverage: 31%)
Pfam:
InterPro:
- Nuclear factor of activated T cells (NFAT)
- Rel homology domain, DNA-binding domain
- Rel homology domain (RHD), DNA-binding domain superfamily
- p53-like transcription factor, DNA-binding domain superfamily
- Immunoglobulin-like fold
- IPT domain
- Immunoglobulin E-set
- Rel homology dimerisation domain
SCOP:
Chain: F
Length: 56 amino acids
Theoretical weight: 6.73 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: bZIP transcription factor
InterPro:
CATH: Single alpha-helices involved in coiled-coils or other helix-helix interfaces
SCOP: Leucine zipper domain
Length: 56 amino acids
Theoretical weight: 6.73 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P01100 (Residues: 138-193; Coverage: 15%)
P01100 (Residues: 138-193; Coverage: 15%)
Pfam: bZIP transcription factor
InterPro:
CATH: Single alpha-helices involved in coiled-coils or other helix-helix interfaces
SCOP: Leucine zipper domain
Chain: J
Length: 56 amino acids
Theoretical weight: 6.58 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: bZIP transcription factor
InterPro:
CATH: Single alpha-helices involved in coiled-coils or other helix-helix interfaces
SCOP: Leucine zipper domain
Length: 56 amino acids
Theoretical weight: 6.58 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P05412 (Residues: 253-308; Coverage: 17%)
P05412 (Residues: 253-308; Coverage: 17%)
Pfam: bZIP transcription factor
InterPro:
CATH: Single alpha-helices involved in coiled-coils or other helix-helix interfaces
SCOP: Leucine zipper domain
Name:
DNA (5'-D(*DTP*DTP*DGP*DGP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DGP*DTP*DTP*DTP*DCP*DAP*DTP*DAP*DG)-3')
Representative chains: A
Length: 20 nucleotides
Theoretical weight: 6.18 KDa
Representative chains: A
Length: 20 nucleotides
Theoretical weight: 6.18 KDa
Name:
DNA (5'-D(*DAP*DAP*DCP*DTP*DAP*DTP*DGP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DCP*DC)-3')
Representative chains: B
Length: 20 nucleotides
Theoretical weight: 6.08 KDa
Representative chains: B
Length: 20 nucleotides
Theoretical weight: 6.08 KDa