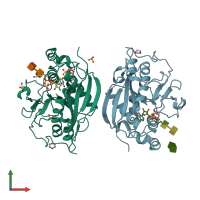Function and Biology Details
Reactions catalysed:
UDP-alpha-D-galactose + D-glucose = UDP + lactose
UDP-alpha-D-galactose + N-acetyl-beta-D-glucosaminyl-(1->3)-beta-D-galactosyl-(1->4)-beta-D-glucosyl-(1<->1)-ceramide = UDP + beta-D-galactosyl-(1->4)-N-acetyl-beta-D-glucosaminyl-(1->3)-beta-D-galactosyl-(1->4)-beta-D-glucosyl-(1<->1)-ceramide
UDP-alpha-D-galactose + N-acetyl-beta-D-glucosaminylglycopeptide = UDP + beta-D-galactosyl-(1->4)-N-acetyl-beta-D-glucosaminylglycopeptide
UDP-alpha-D-galactose + N-acetyl-D-glucosamine = UDP + N-acetyllactosamine
Biochemical function:
Biological process:
Cellular component:
- not assigned
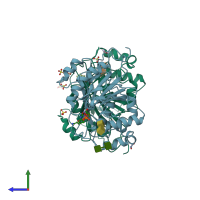
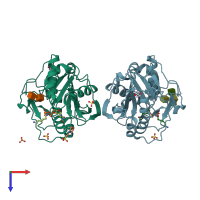
Structure analysis Details
Assembly composition:
monomeric (preferred)
Assembly name:
Beta-1,4-galactosyltransferase 1 (preferred)
PDBe Complex ID:
PDB-CPX-139892 (preferred)
Entry contents:
1 distinct polypeptide molecule
Macromolecules (2 distinct):